LungInsight Data Management Plan*
*DMP designed using “DMP Template for Preclinical Studies”, Blueprint Translational Research Group, Available at: https://journalologytraining.ca/dmp-tools/
1. Data description and collection
1a. Describe the study for which the data are being collected.
Acute respiratory distress syndrome (ARDS) is a severe clinical condition that is investigated experimentally using preclinical models of Acute Lung Injury (ALI). Histological assessment in these models is critical for understanding disease mechanisms and evaluating potential interventions. However, current scoring methods are time-consuming, susceptible to selection bias, and limited by human interpretation, which can undermine the accuracy and reproducibility of findings. To address these challenges, we propose an AI-driven platform to automate histological assessment of ALI. By streamlining analysis, reducing costs, minimizing bias, and improving reproducibility, this tool will provide a robust and scalable foundation for advancing preclinical lung injury research and accelerating the development of new therapies.
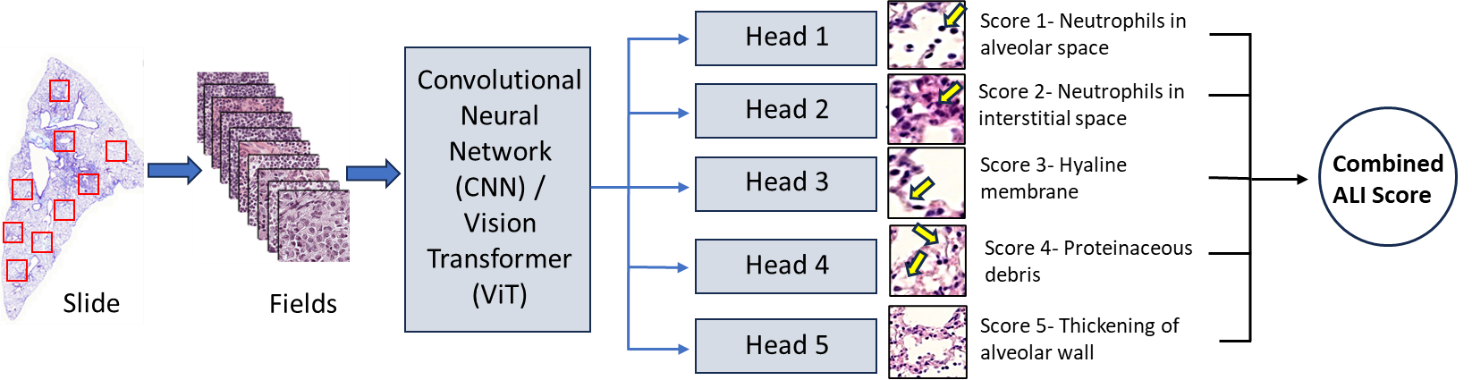
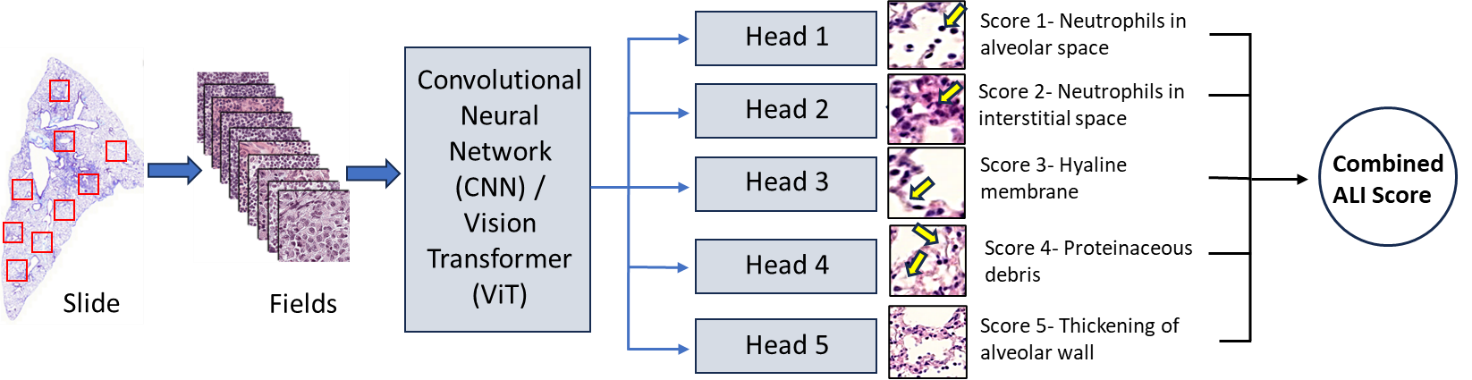
An international team of lung injury experts will contribute histology samples from their laboratories, generated from established ALI animal models using standardized staining protocols. Whole‑slide images (WSIs) will be processed into standardized, fixed‑size tiles to ensure harmonization of image inputs across sites. A random subset of these tiles will be selected for expert annotation by highly qualified personnel (HQP, research staff and senior trainees) using the LungInsightAnnotation application, a cloud-based platform, labeling key injury features using accepted histological scoring criteria. The tool, implemented in Python and Docker, enables asynchronous annotation of image tiles, with all data securely stored in the cloud. To reduce annotator workload and minimize inter-observer variability, the platform generates preliminary region-level annotations by identifying features using classical computer vision techniques based on morphological characteristics. In addition, it provides real-time feedback on annotation consensus, thereby further mitigating inter-observer variability. These annotated tiles will form the training dataset for deep learning models developed in Python, including convolutional neural networks (CNNs) and Vision Transformers (ViTs), enabling automated detection and quantification of lung injury severity. Model performance will be refined through iterative testing and cross-validation, then evaluated on a completely external test set (external animal model and laboratories) to ensure unbiased assessment.
Our specific objectives are to:
-
- Develop an open-source AI-based software for analyzing ALI histology, and
-
- Validate its performance using external datasets.
1b. What types of data will you collect, create, link to, acquire and/or record?
Some pre-analytical metadata for previously prepared or archived histology slides (e.g., microtome blade model/lot) are unrecoverable as this is not commonly recorded information. These fields will remain in our schema and, for all newly accessioned slides, will be prospectively captured.
Animal and housing
| What data is being collected | Description | The type of data being collected |
|---|---|---|
| Type of animal model | Acute lung injury (ALI) model | Text (Nominal) |
| Animal species | e.g. mouse, rat, hamster, pig | |
| Animal Strain | e.g. C57BL/6, Sprague-Dawley | Text (Nominal) |
| Vendor | e.g. Charles River | Text (Nominal) |
| Age | Numeric (Discrete) | |
| Biological Sex | Male or female | Text (Nominal) |
| Genetic modifications | e.g. knock out/in genes, transgenic. And list the target genes | Text (Nominal) |
| Body Weight | In grams | Numeric (continuous) |
| Enrichment materials | e.g. nesting material, tunnels | Text (Nominal) |
| Food | e.g. standard diet, high-fat diet | Text (Nominal) |
| Water type | e.g. medicated water, RO, chlorinated | Text (Nominal) |
| Light/dark cycle | e.g. 12 hrs of light and 12 hrs of dark | Text (Nominal) |
Lung Injury Model
| What data is being collected | Description | The type of data being collected |
|---|---|---|
| Induction method | LPS, acid aspiration, hemorrhagic shock | Text (Nominal) |
| Co-morbidities | e.g. diabetes | Text (Nominal) |
| Duration of injury | In hours | Numeric (continuous) |
| Animal wellness scores at endpoint | Numeric (continuous) | |
| Measure of ALI severity at endpoint | Numeric (continuous) | |
| Intervention (if applicable) | e.g. antibiotics | Text (Nominal) |
| Concentration of intervention (if applicable) | Numeric (discrete) | |
| Time of administration of the intervention (if applicable) | e.g. 24hrs after disease induction | Numeric (discrete) |
| Route of administration of the intervention (if applicable) | e.g. intravenous | Text (Nominal) |
| Duration the intervention is being applied for | Numeric (discrete) | |
| Ventilator model# (if applicable) | Numeric (discrete) | |
| Ventilator settings (if applicable) | Text (Nominal) | |
| Total bronchoalveolar protein concentration | In mg/mL | Numeric (discrete) |
| Number of neutrophils in the bronchoalveolar fluid | Numeric (discrete) | |
| Concentration of proinflammatory cytokines in the bronchoalveolar fluid | e.g. IL-6 concentration | Numeric (discrete) |
Histology slide preparation data/metadata
| What data is being collected | Description | The type of data being collected |
|---|---|---|
| Which laboratory does the slide originate from | Text (Nominal) | |
| Which experimental group does this slide belong to | e.g. control, treated, untreated | Text (Nominal) |
| Lung region sampled | e.g. upper/middle/lower lobe | Text (Nominal) |
| Date animal was sacrificed | (DD-MM-YYYY) | Numeric (discrete) |
| Euthanasia method | e.g. thoracotomy | Text (Nominal) |
| What solution(s) were he tissue stored in | e.g. 70% ethanol | Text (Nominal) |
| What was the storage temperature of the tissue | In Celsius (e.g. room temperature, -4oC, -80oC) | Numeric (discrete) |
| How long was the tissue stored for | e.g. 3 months | Numeric (discrete) |
| Slide preparation date | (DD-MM-YYYY) | Numeric (discrete) |
| EMBEDDING DATA/METADATA | ||
| Fixation method | e.g. formalin (4% PFA) | Text (Nominal) |
| HQP that is preparing the embedded block | Text (Nominal) | |
| Years of experience the HQP has in preparing the block | Numeric (discrete) | |
| Embedding orientation | e.g. ventral side down in the cassette | Text (Nominal) |
| What was the temperature of the embedding medium bath the tissue was submerged in | In Celsius | Numeric (discrete) |
| Embedding medium | e.g. paraffin wax | Text (Nominal) |
| Company tissue processor was purchased from | Leica | Text (Nominal) |
| Tissue processor make and model | e.g. HistoCore PELORIS 3 | Numeric (discrete) |
| Company tissue embedder was purchased from | Leica | Text (Nominal) |
| Tissue embedder make and model | e.g. HistoCore Arcadia | Numeric (discrete) |
| SECTIONING DATA/METADATA | ||
| HQP that is sectioning the block | Text (Nominal) | |
| Years of experience the HQP has in sectioning | Numeric (discrete) | |
| Company microtome was purchased from | Leica | Text (Nominal) |
| Microtome make/model | e.g. HistoCore AUTOCUT | Numeric (discrete) |
| Microtome blade material | e.g. glass, metal, or diamond rock | Text (Nominal) |
| Microtome blade profile | e.g. plano-concave, biconcave, wedge-shaped | Text (Nominal) |
| Microtome blade type | e.g. rotary, sledge, vibrating | Text (Nominal) |
| Temperature of floatation bath | In Celsius | Numeric (discrete) |
| Microtome blade angle | In degrees | Numeric (discrete) |
| Section thickness | In micrometers | Numeric (discrete) |
| Coverslip type | e.g. glass or plastic | Text (Nominal) |
| Coverslip shape | e.g. square, round | Text (Nominal) |
| Coverslip thickness | In millimetres | Numeric (discrete) |
| Coverslip baking conditions (if applicable) |
Temperature: 60oC Time: 1 hour |
Numeric (discrete) |
| STAINING DATA/METADATA | ||
| HQP staining the slide | Text (Nominal) | |
| Years of experience the HQP has in staining slides | Numeric (discrete) | |
| Histological staining dyes | e.g. H&E stain | Text (Nominal) |
| Staining dye catalog# | Abcam cat# ab245880 | Numeric (discrete) |
| Staining dye lot# | Numeric (discrete) | |
| Antibodies if IHC/IF was done | Text (Nominal) | |
| Storage conditions of slides | e.g. room temperature or 4oC | Numeric (discrete) |
| Staining batch ID (if relevant) | Numeric (discrete) | |
Image acquisition
| What data is being collected | Description | The type of data being collected |
| HQP taking the image | Text (Nominal) | |
| Years of experience the HQP has in taking images | Numeric (discrete) | |
| Microscope make/model | e.g. Zeiss AXIO Imager.Z2 Fluorescence Motorized LED Microscope | Text (Nominal) |
| Scanner make/model | e.g. Leica Biosystems Aperio series | Text (Nominal) |
| Objective magnification | e.g. 40X | Numeric (discrete) |
| Scanner mode | e.g. 20x/40x | Numeric (discrete) |
| Image Resolution | In pixels/mm | Numeric (discrete) |
| Modality | e.g. brightfield, fluorescence, polarized | Text (Nominal) |
| Acquisition type | e.g. single FOV, tile scan, z-stack | Text (Nominal) |
| Imaging sample strategy | e.g. ROI, systematic-uniform random | Text (Nominal) |
| Light source type | e.g. LED, halogen, laser | Text (Nominal) |
| Imaging software | e.g. Aperio ImageScope | Text (Nominal) |
| Imaging software version | e.g. v12.3.3 | Text (Nominal) |
| File format of image | e.g. .png | Text (Nominal) |
Tile scoring
| What data is being collected | Description | The type of data being collected |
| HQP doing the scoring | ||
| Years of experience this HQP has in scoring | ||
| Tile size | In micrometers | Numeric (discrete) |
| Tile coordinates | Numeric (discrete) | |
| Overlap % | Numeric (discrete) | |
| stitching algorithm/version | Text (Nominal) | |
| Flat-field correction | Text (Nominal) | |
| Quality control (QC) pass or fail flag | Does the image pass the QC test | Text (Nominal) |
| Exclusion criteria | Reasoning why the image failed QC | Text (Nominal) |
| Absolute number of Neutrophils in the alveolar space | Numeric (discrete) | |
| Absolute number of Neutrophils in the interstitial space | Numeric (discrete) | |
| Absolute number of Hyaline membranes | Numeric (discrete) | |
| Absolute number of Proteinaceous debris filling the airspaces | Numeric (discrete) | |
| Measuring the alveolar septal thickening | In nanometers | Numeric (discrete) |
| Neutrophils in the alveolar space |
Scoring system: No Neutrophils: the score is 0 1-5 Neutrophils: the score is 1 >5 Neutrophils: the score is 2 |
Numeric (discrete) |
| Neutrophils in the interstitial space |
Scoring system: No Neutrophils: the score is 0 1-5 Neutrophils: the score is 1 >5 Neutrophils: the score is 2 |
Numeric (discrete) |
| Hyaline membranes |
Scoring system: No membrane: the score is 0 1 membrane: the score is 1 >1 membrane: the score is 2 |
Numeric (discrete) |
| Proteinaceous debris filling the airspaces |
Scoring system: No debris: the score is 0 1 debris: the score is 1 >1 debris: the score is 2 |
Numeric (discrete) |
| Alveolar septal thickening |
Scoring system: <2x thickness: the score is 0 2x – 4x thickness: the score is 1 >4X thickness: the score is 2 |
Numeric (discrete) |
AI-specific metadata
| What data is being collected | Description | The type of data being collected |
| Record colour normalization method | e.g. Reinhard, Macenko, Vahadane | Text (Nominal) |
| Image augmentations | e.g. rotations/flips, color jitter, Gaussian blur | Text (Nominal) |
| Which tiles were used for Self-supervised pretraining | Text (Nominal) | |
| How were these tiles chosen for Self-supervised pretraining | e.g. random, criteria for exclusion of artifacts | Text (Nominal) |
| Hyperparameter tuning metrics | Score-prediction metrics (MSE, spearman), detection metrics (Accuracy, F1, Precision, Recall, AUROC) | Text (Nominal) |
| Minimum specs needed to run the AI | e.g. GPU type, VRAM, CPU, RAM, CUDA version | Text (Nominal) |
| Training configuration | Key hyperpatameters (batch size, learning rate, optimizer, loss function, epochs, random seeds) | Text (Nominal) |
| Training dataset references | Description of the training/validation sets; number of images/fields | Text (Nominal) |
| Model format and size | e.g. .onnx, .h, .pth with the total size in MB | Text (Nominal) |
| Model version identifier | Git commit hash of trained model | Text (Nominal) |
1c. How will new data be collected or produced and/or how will existing data be re-used?
This study will exclusively collect lung histology slides and their corresponding whole-slide images (WSIs) from collaborating laboratories. The ALI models utilized to develop these images, span a wide range induction method (e.g. LPS, bacterial, viral). While the lungs must be pressure-fixed, sectioned longitudinal, and the slides scanned under standard conditions (40× objective, NA 0.75, ~80% compression, 168-bit color). Partner sites will upload their WSIs to a centralized cloud storage and the images will be converted to the accepted formats (MRXS, SVS, NDPI, DICOM/OME-TIFF).
To generate the tiles used for scoring, the images scanned under standard conditions and stain-normalized using the Reinhard method, were first divided into fields with tissue segmentation to exclude non-informative regions (e.g., torn tissues, edge artifacts). While gradient boosting was used to identify the background and blurry areas. The remaining fields are then saved as standard 16-bit RGB image formats (e.g., SVS, TIFF, JPG, PNG) and randomly assigned to two annotators for scoring.
The primary data being generated is the absolute number of each parameter used in the lung injury score and the injury score itself from both the annotators and the AI model. The score is based off this system:
| Parameter | Score per field | ||
| 0 | 1 | 2 | |
| Neutrophils in the alveolar space | None | 1 – 5 | >5 |
| Neutrophils in the interstitial space | None | 1 – 5 | >5 |
| Hyaline memebrane | None | 1 | >1 |
| Proteinaceous debris filling the airspaces | None | 1 | >1 |
| Alveolar septal thickening | <2X | 2X – 4X | >4X |
1d. What file formats will your data be collected in? Will these formats allow for data reuse, sharing, and long-term access to the data?
Lung histology WSIs will be uploaded to a centralized cloud storage and the images will be converted to the accepted formats (MRXS, SVS, NDPI, DICOM/OME-TIFF). The fields used for scoring will be saved as standard 16-bit RGB image formats (e.g., SVS, TIFF, JPG, PNG), which is a format allowing for reuse, sharing, and long-term access.
1e. What conventions and procedures will you use to structure, name and version-control your files to help you and others better understand how your data are organized?
We have standardized the file naming convention to capture all necessary information without compromising any implemented blinding protocols. The file name, as shown in Figure 1, will be composed of four sections, outlined as follows.
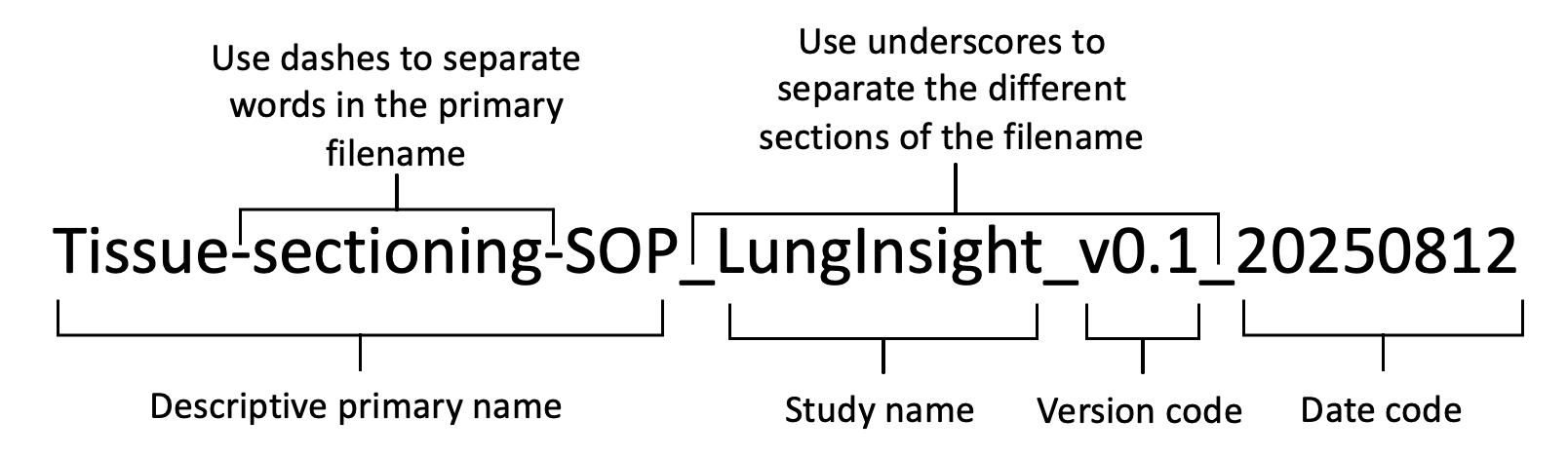
The descriptive primary name will be short (≤25 characters) but meaningfully describe the contents of the file. It should not contain spaces. Rather than using spaces, the use of dashes should distinguish different words within the primary file name.
The study shorthand name should not contain any spaces.
The study short name for this project is “LungInsight”
The date code will reflect the day the data file was generated. We will use ISO 86011 format: YYYY-MM-DD.
The version code will be used to distinguish different versions of the document.
For example, lung-dissection-sop_LungInsight_v0.3_20250812
2. Documentation and Metadata
2a. What documentation will be needed for the data to be read and interpreted correctly in the future?
To help future researchers interpret our published data we will include the following:
-
- Experimental SOPs
- Definitions of preclinical characteristics and outcomes
- Naming conventions
- We will also include a list of all personnel involved in the project, along with a list of their tasks throughout the project.
- Description of the scoring system
- Source code (completely annotated) for the LungInsightScore and LungInsightAnnotation Software README files to execute the code
- Trained AI models (Weights) in formats such as ONNX and HDF5.
- Hyperparameters and training settings for each AI model, provided as a concise config file and brief summary for reproducibility.
2b. How will you make sure that documentation is created and captured consistently throughout your project?
To ensure accuracy, consistency, and completeness, we will institute the following measures:
-
- SOPs will be reviewed with all involved personnel prior to experiments.
- To ensure consistent lung injury scoring online learning modules for each parameter that will be openly accessible on www.LungInsight.ai.
- Prior to beginning “live” field annotation, all annotators will be required to pass modules as well as standardized testing for each parameter
- Every field will undergo duplicate assessment by two individuals
- LungInsightAnnotation will store all scoring data (e.g. neutrophil count, coordinates) in their respective standardized format
- Slides will be converted to MRXS, SVS, NDPI, DICOM/OME-TIFF formats
- Tiles will be converted to standard image formats (e.g., SVS, TIFF, JPG, PNG)
- All normalization techniques and augmentations performed will be recorded for each image
2c. If you are using a metadata standard and/or tools to document and describe your data, please list here.
We will standardize our biological/medical related vocabulary based off the Darwin Core: https://www.tdwg.org/standards/dwc/
We will standardize AI related vocabulary based off the ISO/IEC 22989:2022 – Information technology – Artificial intelligence – Artificial intelligence concepts and terminology (https://www.iso.org/standard/74296.html)
Since we will be uploading our data to Open Science Framework (https://osf.io/), we will follow their metadata standards, which states to follow a metadata scheme that is common to our project. Therefore, we will follow the metadata scheme developed by DataCite: https://schema.datacite.org/
3. Storage and Backup
3a. What are the anticipated storage requirements for your project, in terms of storage space (in megabytes, gigabytes, terabytes, etc.) and the length of time you will be storing it?
The estimated storage-space is 1 terabyte of data. There are no restrictions on how long to retain the data as we will generate non-sensitive laboratory animal data.
3b. How and where will your data be stored and backed up during your research project?
The data will be stored using the 3-2-1 backup rule. Three copies of every piece of data will be generated, the original data and two backups. During the data acquisition stage, all data generated from different experiments will be stored on the personal computer of the person collecting the data and stored in their own project folder. This project folder must be linked to the cloud-based Corporate Microsoft 365 platform, SharePoint, which will synchronize to our lab computer.
The two backup copies of this data will be stored on two different types of media, one on a SharePoint that is dedicated to this project and one on an external hard drive owned by the PI.
The dedicated lab computer is not assigned to one individual but is controlled by the PI (password-protected). The data will be backed up on a SharePoint dedicated to this project accessible only by the PI, students, and staff members directly involved.
The external hard drive will be stored by the PI and will be updated every three months to add any new versions created since the last update.
3c. How will the research team and other collaborators access, modify, and contribute data throughout the project?
During this research project, all data will be stored in SharePoint cloud storage that will be shared between our PI, students, and staff members directly involved. The PI will need to grant access to others not directly involved in the project, in which they will have to log in (username and password). All personnel directly involved in the project will be able to modify and process the data. For any data modification, a new version file will be generated and reflected in the file name (see file naming conventions). Once the raw data is uploaded into the SharePoint (non-editable version), a copy of the data will be made for modifications (editable version). Furthermore, modification is restricted to the data processing, and not modifications to the original raw data. For any modification done to data, a new file will be generated with its file name indicating which version we are on.
4. Preservation
4a. Where will you deposit your data for long-term preservation and access at the end of your research project?
Upon completion of the project and publication in a peer-reviewed journal, original data, metadata, and the standard operating procedures (SOPs) will be made publicly available on the Open Science Framework in their respective formats (csv,.txt, avi). Slide images will be saved as 16-bit RGB TIFF great for long term preservation. Due their large size, we will house original images and the AI models in the Federated Research Data Repository.
As for the source code, all codes will be available on GitHub and accompanied with “READ ME” files containing detailed instructions on installation, dependencies, and execution. The code will remain publicly accessible to ensure long-term preservation and ease of reuse.
4b. Indicate how you will ensure your data is preservation ready. Consider preservation-friendly file formats, ensuring file integrity, anonymization and de-identification, inclusion of supporting documentation.
Along with uploading files in their original format, non-proprietary, preservation friendly, file formats will be used. Data will be saved as .csv, images will be saved as 16-bit RGB TIFF, any text files (e.g. SOPs) will be saved as .txt.
The source code will be saved in UTF-8 encoded plain text that is non-proprietary and preservation friendly.
The trained AI models (CNN and Vision Transformer) will be preserved and shared in open, interoperable formats. Model weights will be exported in preservation-friendly formats such as ONNX and HDF5, with configuration files provided separately in concise JSON/YAML.
5. Sharing and Reuse
5a. What data will you be sharing and in what form? (e.g. raw, processed, analyzed, final, and metadata).
Once this study is completed, all data forms (raw, processed, analyzed, final, and metadata) will be published in peer-reviewed journals. The data stored in the Open Science Framework will be linked to the published article via DOI. The final analyzed data will also be uploaded to Open Science Framework in their respective preservation friendly format (.csv, .txt, 16-bit RGB TIFF, .avi). Given that our imaging data and trained AI models will exceed the 50GB capacity of Open Science Framework, we will house original images and the AI models in the Federated Research Data Repository (https://www.frdr-dfdr.ca/repo/) and the source code on GitHub. We will clearly link to this data from our project on the Open Science Framework. The required protocols, SOPs, and scoring lists to process the raw data to generate the final analyzed data will be included. We will also provide metadata, with a readme file with the coding, variables, naming conventions, and standards.
5b. What type of end-user will you use for your data?
We will share all materials via a Creative Commons license (CC BY 4.0.). The data generated is not sensitive (i.e. all lab animal data) and therefore, a CC license is sufficient. A CC BY license enables users to modify and redistribute data in any form, with proper credit given to our research group (i.e. the original generators of the data).
5c. What steps will be taken to help the research community know that your data exists?
To make our data findable and accessible, the data and metadata will be archived and shared via the Open Science Framework. The DOI number provided by the Open Science Framework will be included in the publication. DOIs promote academic credit, direct citation, and tangible metrics that our group will track. The DOI will also link to the final publication(s), as well as information on study funders and our institute where the study was performed. In addition to publishing our data in a peer-reviewed journal, we will promote our research via conference presentations, and poster presentations. The ORCID ID of every researcher involved will be linked in the publication.
In addition to uploading our data into Open Science Framework, we have also created a website, www.LungInsight.ai, that will notify researchers on any updates to our project via newsletters and publications.
To make the data interoperable, we will share detailed metadata (workflows, vocabularies, processes, and standards) and the data will be shared in preservation friendly formats as detailed above. To make the data will share it under a Creative Commons CC-BY-4.0 license.
6. Responsibilities and Resources
6a. Identify who will be responsible for managing this project’s data during and after the project and the major data management tasks for which they will be responsible.
This Team will consist of
| Individual | Position | Role | |
|---|---|---|---|
| Dr. Arvid Mer | NPA | Investigator | |
| Dr. Majid Komeili | Co-principal applicant | Investigator | |
| Dr. Manoj Lalu | Co-principal applicant | Investigator | |
| Dr. Dean Fergusson | Co-applicant | Investigator | |
| Dr. Arya Rahgozar | Collaborator | ||
| Dr. Sean Gil | Collaborator | Sharing ALI histology slides | |
| Dr. Haibo Zhang | Collaborator | Sharing ALI histology slides | |
| Dr. Arnold Kristof | Collaborator | Sharing ALI histology slides | |
| Dr. Bernard Thebaud | Collaborator | Sharing ALI histology slides | |
| Dr. Claudia DosSantos | Collaborator | Sharing ALI histology slides | |
| Dr. Christian Lehmann | Collaborator | Sharing ALI histology slides | |
| Dr. Duncan Stewart | Collaborator | Sharing ALI histology slides | |
| Dr. Braedon McDonald | Collaborator | Sharing ALI histology slides | |
| Dr. Katey Rayner | Collaborator | Sharing ALI histology slides | |
| Dr. Eric Schmidt | Collaborator | Sharing ALI histology slides | |
| Dr. Julie Bastarache | Collaborator | Sharing ALI histology slides | |
| Dr. Patrica Rocco | Collaborator | Sharing ALI histology slides | |
| Dr. Forough Jahandideh | Research Associate | Project manager | |
| Eva Kuhar | HQP | Annotator, managing ALI slides data and metadata | |
| Zoe Fisk | HQP | Annotator, managing ALI slides data and metadata | |
| Amir Ebrahimi | HQP | AI model development, Managing AI data and metadata | |
| MohammadReza Zarei | HQP | AI explainability and implementation, Managing AI data and metadata |
The names and specific roles of highly qualified personnel (HQP) will be added to this DMP as they are onboarded to the project.
6b. How will responsibilities for managing data activities be handled if substantive changes occur in the personnel overseeing the project’s data, including a change of Principal Investigator?
If any staff or trainees leave the project prior to completion, a current staff or trainee will replace them (or new personnel will be hired, if required). New personnel will receive adequate training to ensure competency image processing and lung scoring. Training will be enabled through project specific SOPs as well as videos found on www.LungInsight.ai. Furthermore, the staff/trainee will go through an off-boarding protocol outlining all the data they have generated, update any SOPs created, provide the location materials purchased, location of all the tissue/samples collected, etc
If the project needs to be transferred to a new principal investigator, then all responsibilities outlined in this DMP will be transferred as well.
6c. What resources will you require to implement your data management plan? What do you estimate the overall cost for data management to be?
The costs of implementing this DMP will consistent of:
-
- Hiring a dedicated staff membrane to execute the DMP
- Fees for keeping www.LungInsight.ai running
- Cost of the hard drives to store the images
For data sharing, since we are using the Open Science Framework, there is no cost for uploading our data. Given that our imaging data and AI models will exceed the 50GB capacity of Open Science Framework, we will house original images in the Federated Research Data Repository. We will clearly link to this data from our project on the Open Science Framework.
7. Ethics and Legal Compliance
7a. If your research project includes sensitive data, how will you ensure that it is securely managed and accessible only to approved members of the project?
Not applicable as only non-sensitive laboratory animal data will be collected/generated in our study.
7b. If applicable, what strategies will you undertake to address secondary uses of sensitive data?
Not applicable as the ‘participants’ are lung histology images.
7c. How will you manage legal, ethical, and intellectual property issues?
To ensure discoverability and access, we will deposit all datasets and accompanying metadata on the Open Science Framework (OSF). Once uploaded, we will receive a Digital Object Identifier (DOI), which we will cross-reference with publications, funding acknowledgements, and our institutional affiliation. In parallel, we will disseminate results through conference talks and poster sessions. Beyond the OSF archive, we will maintain a project website (www.LungInsight.ai) to announce updates—such as newsletters and publications—to the research community. To support interoperability, we will publish rich, machine-readable metadata documenting workflows, controlled vocabularies, processes, and standards, and we will share files in preservation-friendly formats as specified above. All materials will be released under a Creative Commons Attribution 4.0 (CC BY 4.0) license.
Version History
| DMP version number | Date Issued | Summary of Revisions |
|---|---|---|
| DMP_LungInsight_v0.1_20250812 | 15-08-2025 | Initial draft by Brian Dorus; with comments provided by Amir Ebrahimi, Eva Kuhar, and Manoj Lalu |
| DMP_LungInsight_v0.2_20251006 | 06-10-2025 | Initial comments addressed by Brian Dorus; second round of comments provided by Manoj Lalu |
| DMP_LungInsight_v0.3_20251007 | 07-10-2025 | Second round of comments addressed by Brian Dorus |
| DMP_LungInsight_v0.4_20251007 | 07-10-2025 | Comments provided by Forough Jahandideh and Zoe Fisk, and addressed by Brian Dorus |
| DMP_LungInsight_v0.5_20251009 | 09-10-2025 | Comments provided by Arvind Mer, Majid Komeili, Amir Ebrahimi, and Mohammad Reza Zarei and addressed by Brian Dorus |
| DMP_LungInsight_v0.6_20251010 | 10-10-2025 |
LungInsight Data Management Plan*
*DMP designed using “DMP Template for Preclinical Studies”, Blueprint Translational Research Group, Available at: https://journalologytraining.ca/dmp-tools/
1. Data description and collection
1a. Describe the study for which the data are being collected.
Acute respiratory distress syndrome (ARDS) is a severe clinical condition that is investigated experimentally using preclinical models of Acute Lung Injury (ALI). Histological assessment in these models is critical for understanding disease mechanisms and evaluating potential interventions. However, current scoring methods are time-consuming, susceptible to selection bias, and limited by human interpretation, which can undermine the accuracy and reproducibility of findings. To address these challenges, we propose an AI-driven platform to automate histological assessment of ALI. By streamlining analysis, reducing costs, minimizing bias, and improving reproducibility, this tool will provide a robust and scalable foundation for advancing preclinical lung injury research and accelerating the development of new therapies.
An international team of lung injury experts will contribute histology samples from their laboratories, generated from established ALI animal models using standardized staining protocols. Whole‑slide images (WSIs) will be processed into standardized, fixed‑size tiles to ensure harmonization of image inputs across sites. A representative subset of these tiles (typically 5–15 fields, average 10) will be selected for expert annotation by highly qualified personnel (HQP, research staff and senior trainees) using the LungInsightAnnotation application, a cloud-based platform, labeling key injury features using accepted histological scoring criteria. The tool, implemented in Python and Docker, enables asynchronous annotation of image tiles, with all data securely stored in the cloud. To reduce annotator workload and minimize inter-observer variability, the platform generates preliminary region-level annotations by identifying features using classical computer vision techniques based on morphological characteristics. In addition, it provides real-time feedback on annotation consensus, thereby further mitigating inter-observer variability. These annotated tiles will form the training dataset for deep learning models developed in Python, including convolutional neural networks (CNNs) and Vision Transformers (ViTs), enabling automated detection and quantification of lung injury severity. Model performance will be refined through iterative testing and cross-validation, then evaluated on a completely external test set (external animal model and laboratories) to ensure unbiased assessment.
Our specific objectives are to:
- Develop an open-source AI-based software for analyzing ALI histology, and
- Validate its performance using external datasets and analyze generalizability
1b. What types of data will you collect, create, link to, acquire and/or record?
Some pre-analytical metadata for previously prepared or archived histology slides (e.g., microtome blade model/lot) are unrecoverable as this is not commonly recorded information. These fields will remain in our schema and, for all newly accessioned slides, will be prospectively captured.
Animal and housing
|
What data is being collected |
Description |
The type of data being collected |
|
Type of animal model |
Acute lung injury (ALI) |
Text (Nominal) |
|
Animal species |
e.g., mouse, rat, hamster, pig |
|
|
Animal strain |
e.g., C57BL/6, Sprague-Dawley |
Text (Nominal) |
|
Vendor |
e.g., Charles River |
Text (Nominal) |
|
Age |
Numeric (Discrete) |
|
|
Biological sex |
Male or female |
Text (Nominal) |
|
Genetic modifications |
e.g., knock out/in genes, transgenic (and list the target genes) |
Text (Nominal) |
|
Body weight |
In grams |
Numeric (continuous) |
|
Enrichment materials |
e.g., nesting material, dome, hut, cylinder |
Text (Nominal) |
|
Food |
e.g., standard diet, high-fat diet |
Text (Nominal) |
|
Housing conditions |
e.g., single or grouped |
Text (nominal) |
|
Light/dark cycle |
e.g., 12 hrs of light and 12 hrs of dark |
Text (Nominal) |
Lung Injury Model
|
What data is being collected |
Description |
The type of data being collected |
|
Induction method |
LPS, acid aspiration, hemorrhagic shock, bacterial, viral |
Text (Nominal) |
|
Co-morbidities |
e.g., diabetes |
Text (Nominal) |
|
Duration of injury |
In hours |
Numeric (continuous) |
|
Animal wellness scores at endpoint |
Numeric (continuous) |
|
|
Measure of ALI severity at endpoint |
Numeric (continuous) |
|
|
Intervention (if applicable) |
e.g., antibiotics |
Text (Nominal) |
|
Concentration of intervention (if applicable) |
Numeric (discrete) |
|
|
Time of intervention administration (if applicable) |
e.g., 24hrs after disease induction |
Numeric (discrete) |
|
Route of intervention administration (if applicable) |
e.g., intravenous |
Text (Nominal) |
|
Duration the intervention is being applied for (if applicable) |
Numeric (discrete) |
|
|
Ventilator model # (if applicable) |
Numeric (discrete) |
|
|
Ventilator settings (if applicable) |
Text (Nominal) |
|
|
Total bronchoalveolar protein concentration |
In mg/mL |
Numeric (discrete) |
|
Number of neutrophils in the bronchoalveolar fluid |
Numeric (discrete) |
|
|
Concentration of proinflammatory cytokines in the bronchoalveolar fluid at a specified timepoint |
e.g., IL-6 concentration |
Numeric (discrete) |
Histology slide preparation data/metadata
|
What data is being collected |
Description |
The type of data being collected |
|
Which laboratory does the slide originate from |
Text (Nominal) |
|
|
Which experimental group does this slide belong to |
e.g., control, treated, untreated |
Text (Nominal) |
|
Lung region sampled |
e.g., upper/middle/lower lobe |
Text (Nominal) |
|
Date animal was sacrificed |
(DD-MM-YYYY) |
Numeric (discrete) |
|
Euthanasia method |
e.g. Cervical dislocation |
Text (Nominal) |
|
What solution(s) were he tissue stored in |
e.g., 70% ethanol |
Text (Nominal) |
|
What was the storage temperature of the tissue |
In Celsius (e.g. room temperature, -4oC, -80oC) |
Numeric (discrete) |
|
How long was the tissue stored for |
e.g., 3 months |
Numeric (discrete) |
|
Slide preparation date |
(DD-MM-YYYY) |
Numeric (discrete) |
|
EMBEDDING DATA/METADATA |
||
|
Fixation method |
e.g., formalin (4% PFA) |
Text (Nominal) |
|
HQP that is preparing the embedded block |
Text (Nominal) |
|
|
Years of experience the HQP has in preparing the block |
Numeric (discrete) |
|
|
Embedding orientation |
e.g., ventral side down in the cassette |
Text (Nominal) |
|
What was the temperature of the embedding medium bath the tissue was submerged in |
In Celsius |
Numeric (discrete) |
|
Embedding medium |
e.g., paraffin wax |
Text (Nominal) |
|
Company tissue processor was purchased from |
Leica |
Text (Nominal) |
|
Tissue processor make and model |
e.g., HistoCore PELORIS 3 |
Numeric (discrete) |
|
Company tissue embedder was purchased from |
Leica |
Text (Nominal) |
|
Tissue embedder make and model |
e.g., HistoCore Arcadia |
Numeric (discrete) |
|
SECTIONING DATA/METADATA |
||
|
HQP that is sectioning the block |
Text (Nominal) |
|
|
Years of experience the HQP has in sectioning |
Numeric (discrete) |
|
|
Company microtome was purchased from |
Leica |
Text (Nominal) |
|
Microtome make/model |
e.g., HistoCore AUTOCUT |
Numeric (discrete) |
|
Microtome blade material |
e.g., glass, metal, or diamond rock |
Text (Nominal) |
|
Microtome blade profile |
e.g., plano-concave, biconcave, wedge-shaped |
Text (Nominal) |
|
Microtome blade type |
e.g., rotary, sledge, vibrating |
Text (Nominal) |
|
Temperature of floatation bath |
In Celsius |
Numeric (discrete) |
|
Microtome blade angle |
In degrees |
Numeric (discrete) |
|
Section thickness |
In micrometers |
Numeric (discrete) |
|
Coverslip type |
e.g., glass or plastic |
Text (Nominal) |
|
Coverslip shape |
e.g., square, round |
Text (Nominal) |
|
Coverslip thickness |
In millimetres |
Numeric (discrete) |
|
Coverslip baking conditions (if applicable) |
Temperature: 60oC Time: 1 hour |
Numeric (discrete) |
|
STAINING DATA/METADATA |
||
|
HQP staining the slide |
Text (Nominal) |
|
|
Years of experience the HQP has in staining slides |
Numeric (discrete) |
|
|
Histological staining dyes |
e.g., H&E stain |
Text (Nominal) |
|
Staining dye catalog# |
Abcam cat# ab245880 |
Numeric (discrete) |
|
Staining dye lot# |
Numeric (discrete) |
|
|
Antibodies if IHC/IF was done |
Text (Nominal) |
|
|
Storage conditions of slides |
e.g., room temperature or 4oC |
Numeric (discrete) |
|
Staining batch ID (if relevant) |
Numeric (discrete) |
|
Image acquisition
|
What data is being collected |
Description |
The type of data being collected |
|
HQP taking the image |
Text (Nominal) |
|
|
Years of experience the HQP has in taking images |
Numeric (discrete) |
|
|
Microscope make/model |
e.g., Zeiss AXIO Imager.Z2 Fluorescence Motorized LED Microscope |
Text (Nominal) |
|
Scanner make/model |
e.g., Leica Biosystems Aperio series |
Text (Nominal) |
|
Objective magnification |
e.g., 40X |
Numeric (discrete) |
|
Scanner mode |
e.g., 20x/40x |
Numeric (discrete) |
|
Image resolution |
In pixels/mm |
Numeric (discrete) |
|
Modality |
e.g., brightfield, fluorescence, polarized |
Text (Nominal) |
|
Acquisition type |
e.g., single FOV, tile scan, z-stack |
Text (Nominal) |
|
Imaging sample strategy |
e.g., ROI, systematic-uniform random |
Text (Nominal) |
|
Light source type |
e.g., LED, halogen, laser |
Text (Nominal) |
|
Imaging software |
e.g., Aperio ImageScope |
Text (Nominal) |
|
Imaging software version |
e.g., v12.3.3 |
Text (Nominal) |
|
File format of image |
e.g., .png |
Text (Nominal) |
Tile scoring
|
What data is being collected |
Description |
The type of data being collected |
|
HQP doing the scoring |
||
|
Years of experience this HQP has in scoring |
||
|
Tile size |
Area (micrometers2) |
Numeric (discrete) |
|
Tile coordinates |
Numeric (discrete) |
|
|
Overlap % |
Numeric (discrete) |
|
|
Flat-field correction |
Text (Nominal) |
|
|
Quality control (QC) pass or fail flag |
Does the image pass the QC test |
Text (Nominal) |
|
Exclusion criteria |
Reasoning why the image failed QC |
Text (Nominal) |
|
Absolute number of neutrophils in the alveolar space |
Numeric (discrete) |
|
|
Absolute number of neutrophils in the interstitial space |
Numeric (discrete) |
|
|
Absolute number of hyaline membranes |
Numeric (discrete) |
|
|
Absolute number of Proteinaceous debris filling the airspaces |
Numeric (discrete) |
|
|
Measuring the alveolar septal thickening |
In nanometers |
Numeric (discrete) |
|
Neutrophils in the alveolar space |
Scoring system: No Neutrophils: the score is 0 1-5 Neutrophils: the score is 1 >5 Neutrophils: the score is 2 |
Numeric (discrete) |
|
Neutrophils in the interstitial space |
Scoring system: No Neutrophils: the score is 0 1-5 Neutrophils: the score is 1 >5 Neutrophils: the score is 2 |
Numeric (discrete) |
|
Hyaline membranes |
Scoring system: No membrane: the score is 0 1 membrane: the score is 1 >1 membrane: the score is 2 |
Numeric (discrete) |
|
Proteinaceous debris filling the airspaces |
Scoring system: No debris: the score is 0 1 debris: the score is 1 >1 debris: the score is 2 |
Numeric (discrete) |
|
Alveolar septal thickening |
Scoring system: <2x thickness: the score is 0 2x – 4x thickness: the score is 1 >4X thickness: the score is 2 |
Numeric (discrete) |
AI-specific metadata
|
What data is being collected |
Description |
The type of data being collected |
|
Record colour normalization method |
e.g., Reinhard, Macenko, Vahadane |
Text (Nominal) |
|
Image augmentations |
e.g., rotations/flips, color jitter, Gaussian blur |
Text (Nominal) |
|
Which tiles were used for Self-supervised pretraining |
Text (Nominal) |
|
|
How were these tiles chosen for Self-supervised pretraining |
e.g., random, criteria for exclusion of artifacts |
Text (Nominal) |
|
Hyperparameter tuning metrics |
Score-prediction metrics (MSE, spearman), detection metrics (Accuracy, F1, Precision, Recall, AUROC) |
Text (Nominal) |
|
Minimum specs needed to run the AI |
e.g., GPU type, VRAM, CPU, RAM, CUDA version |
Text (Nominal) |
|
Training configuration |
Key hyperpatameters (batch size, learning rate, optimizer, loss function, epochs, random seeds) |
Text (Nominal) |
|
Training dataset references |
Description of the training/validation sets; number of images/fields |
Text (Nominal) |
|
Model format and size |
e.g., .onnx, .h, .pth with the total size in MB |
Text (Nominal) |
|
Model version identifier |
Git commit hash of trained model |
Text (Nominal) |
1c. How will new data be collected or produced and/or how will existing data be re-used?
This study will exclusively collect lung histology slides and their corresponding whole-slide images (WSIs) from collaborating laboratories. The ALI models utilized to develop these images, span a wide range induction method (e.g. LPS, bacterial, viral). While the lungs must be pressure-fixed, sectioned longitudinal, and the slides scanned under standard conditions (40× objective, NA 0.75, ~80% compression, 168-bit color). Partner sites will upload their WSIs to a centralized cloud storage and the images will be converted to the accepted formats (MRXS, SVS, NDPI, DICOM/OME-TIFF).
To generate the tiles for scoring, the images scanned under standard conditions and stain-normalized using the Reinhard method, will be first divided into tiles with tissue segmentation to exclude non-informative regions (e.g., torn tissues, edge artifacts). Gradient boosting will be used to identify the background and blurry areas. The remaining tiles will then be saved as standard 16-bit RGB image formats (e.g., SVS, TIFF, JPG, PNG) and randomly assigned to two annotators for scoring.
The primary data being generated is the absolute number of each parameter used in the lung injury score and the injury score itself from both the annotators and the AI model. The score is based off this system:
|
Parameter |
Score per field |
||
|
0 |
1 |
2 |
|
|
Neutrophils in the alveolar space |
None |
1 – 5 |
>5 |
|
Neutrophils in the interstitial space |
None |
1 – 5 |
>5 |
|
Hyaline memebrane |
None |
1 |
>1 |
|
Proteinaceous debris filling the airspaces |
None |
1 |
>1 |
|
Alveolar septal thickening |
<2X |
2X – 4X |
>4X |
1d. What file formats will your data be collected in? Will these formats allow for data reuse, sharing, and long-term access to the data?
Lung histology WSIs will be uploaded to a centralized cloud storage and the images will be converted to the accepted formats (MRXS, SVS, NDPI, DICOM/OME-TIFF). The fields used for scoring will be saved as standard 16-bit RGB image formats (e.g., SVS, TIFF, JPG, PNG), which is a format allowing for reuse, sharing, and long-term access.
1e. What conventions and procedures will you use to structure, name and version-control your files to help you and others better understand how your data are organized?
We have standardized the file naming convention to capture all necessary information without compromising any implemented blinding protocols. The file name, as shown in Figure 1, will be composed of four sections, outlined as follows.
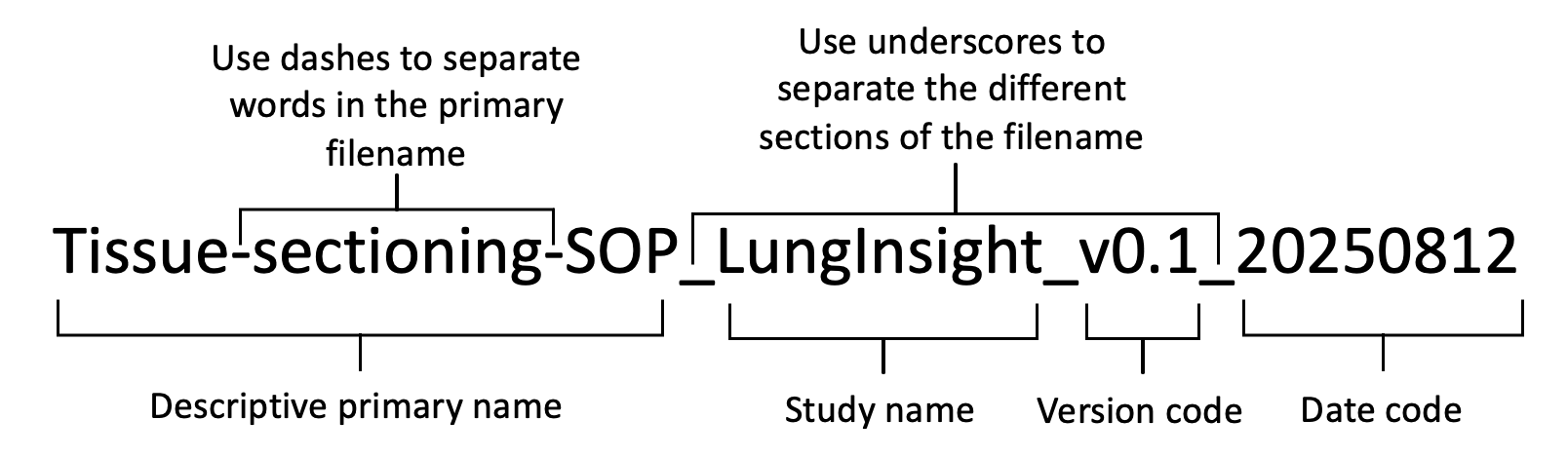
Figure 1. Naming convention for files
The descriptive primary name will be short (≤25 characters) but meaningfully describe the contents of the file. It should not contain spaces. Rather than using spaces, the use of dashes should distinguish different words within the primary file name.
The study shorthand name should not contain any spaces.
The study short name for this project is “LungInsight”
The date code will reflect the day the data file was generated. We will use ISO 86011 format: YYYY-MM-DD.
The version code will be used to distinguish different versions of the document.
For example, lung-dissection-sop_LungInsight_v0.3_20250812
2. Documentation and Metadata
2a. What documentation will be needed for the data to be read and interpreted correctly in the future?
To help future researchers interpret our published data we will include the following:
- Experimental SOPs
- Definitions of preclinical characteristics and outcomes
- Naming conventions
- We will also include a list of all personnel involved in the project, along with a list of their tasks throughout the project.
- Description of the scoring system
- Source code (completely annotated) for the LungInsightScore and LungInsightAnnotation Software README files to execute the code
- Trained AI models (Weights) in formats such as ONNX and HDF5.
- Hyperparameters and training settings for each AI model, provided as a concise config file and brief summary for reproducibility.
2b. How will you make sure that documentation is created and captured consistently throughout your project?
To ensure accuracy, consistency, and completeness, we will institute the following measures:
- SOPs will be reviewed with all involved personnel prior to experiments.
- To ensure consistent lung injury scoring online learning modules for each parameter that will be openly accessible on www.LungInsight.ai.
- Prior to beginning “live” tile annotation, all annotators will be required to pass modules as well as standardized testing for each parameter
- Every tile will undergo duplicate assessment by two individuals
- LungInsightAnnotation will store all scoring data (e.g. neutrophil count, coordinates) in their respective standardized format
- Slides will be converted to MRXS, SVS, NDPI, DICOM/OME-TIFF formats
- Tiles will be converted to standard image formats (e.g., SVS, TIFF, JPG, PNG)
- All normalization techniques and augmentations performed will be recorded for each image
2c. If you are using a metadata standard and/or tools to document and describe your data, please list here.
We will standardize our biological/medical related vocabulary based off the Darwin Core: https://www.tdwg.org/standards/dwc/
We will standardize AI related vocabulary based off the ISO/IEC 22989:2022 – Information technology – Artificial intelligence – Artificial intelligence concepts and terminology (https://www.iso.org/standard/74296.html)
Since we will be uploading our data to Open Science Framework (https://osf.io/), we will follow their metadata standards, which states to follow a metadata scheme that is common to our project. Therefore, we will follow the metadata scheme developed by DataCite: https://schema.datacite.org/
3. Storage and Backup
3a. What are the anticipated storage requirements for your project, in terms of storage space (in megabytes, gigabytes, terabytes, etc.) and the length of time you will be storing it?
The estimated storage-space is 1 terabyte of data. There are no restrictions on how long to retain the data as we will generate non-sensitive laboratory animal data.
3b. How and where will your data be stored and backed up during your research project?
The data will be stored using the 3-2-1 backup rule. Three copies of every piece of data will be generated, the original data and two backups. During the data acquisition stage, all data generated from different experiments will be stored on the personal computer of the person collecting the data and stored in their own project folder. This project folder must be linked to the cloud-based Corporate Microsoft 365 platform, SharePoint, which will synchronize to our lab computer.
The two backup copies of this data will be stored on two different types of media, one on a SharePoint that is dedicated to this project (located on the OHRI’s main server) and one on a complete separated backup server maintained by OHRI.
The dedicated lab computer is not assigned to one individual but is controlled by the PI (password-protected). The data will be backed up on a SharePoint dedicated to this project accessible only by the PI, students, and staff members directly involved.
3c. How will the research team and other collaborators access, modify, and contribute data throughout the project?
During this research project, all data will be stored in SharePoint cloud storage that will be shared between our PI, students, and staff members directly involved. The PI will need to grant access to others not directly involved in the project, in which they will have to log in (username and password). All personnel directly involved in the project will be able to modify and process the data. For any data modification, a new version file will be generated and reflected in the file name (see file naming conventions). Once the raw data is uploaded into the SharePoint (non-editable version), a copy of the data will be made for modifications (editable version). Furthermore, modification is restricted to the data processing, and not modifications to the original raw data. For any modification done to data, a new file will be generated with its file name indicating which version we are on.
4. Preservation
4a. Where will you deposit your data for long-term preservation and access at the end of your research project?
Upon completion of the project and publication in a peer-reviewed journal, original data, metadata, and the standard operating procedures (SOPs) will be made publicly available on the Open Science Framework in their respective formats (csv,.txt, avi). Slide images will be saved as 16-bit RGB TIFF great for long term preservation. Due their large size, we will house original images and the AI models in the Federated Research Data Repository.
As for the source code, all codes will be available on GitHub and accompanied with “READ ME” files containing detailed instructions on installation, dependencies, and execution. The code will remain publicly accessible to ensure long-term preservation and ease of reuse.
4b. Indicate how you will ensure your data is preservation ready. Consider preservation-friendly file formats, ensuring file integrity, anonymization and de-identification, inclusion of supporting documentation.
Along with uploading files in their original format, non-proprietary, preservation friendly, file formats will be used. Data will be saved as .csv, images will be saved as 16-bit RGB TIFF, any text files (e.g., SOPs) will be saved as .txt.
The source code will be saved in UTF-8 encoded plain text that is non-proprietary and preservation friendly.
The trained AI models (CNN and Vision Transformer) will be preserved and shared in open, interoperable formats. Model weights will be exported in preservation-friendly formats such as ONNX and HDF5, with configuration files provided separately in concise JSON/YAML.
5. Sharing and Reuse
5a. What data will you be sharing and in what form? (e.g. raw, processed, analyzed, final, and metadata).
Once this study is completed, all data forms (raw, processed, analyzed, final, and metadata) will be published in peer-reviewed journals. The data stored in the Open Science Framework will be linked to the published article via DOI. The final analyzed data will also be uploaded to Open Science Framework in their respective preservation friendly format (.csv, .txt, 16-bit RGB TIFF, .avi). Given that our imaging data and trained AI models will exceed the 50GB capacity of Open Science Framework, we will house original images and the AI models in the Federated Research Data Repository (https://www.frdr-dfdr.ca/repo/) and the source code on GitHub. We will clearly link to this data from our project on the Open Science Framework. The required protocols, SOPs, and scoring lists to process the raw data to generate the final analyzed data will be included. We will also provide metadata, with a readme file with the coding, variables, naming conventions, and standards.
5b. What type of end-user will you use for your data?
We will share all materials via a Creative Commons license (CC BY 4.0.). The data generated is not sensitive (i.e. all lab animal data) and therefore, a CC license is sufficient. A CC BY license enables users to modify and redistribute data in any form, with proper credit given to our research group (i.e. the original generators of the data).
5c. What steps will be taken to help the research community know that your data exists?
To make our data findable and accessible, the data and metadata will be archived and shared via the Open Science Framework. The DOI number provided by the Open Science Framework will be included in the publication. DOIs promote academic credit, direct citation, and tangible metrics that our group will track. The DOI will also link to the final publication(s), as well as information on study funders and our institute where the study was performed. In addition to publishing our data in a peer-reviewed journal, we will promote our research via conference presentations, and poster presentations. The ORCID ID of every researcher involved will be linked in the publication.
In addition to uploading our data into Open Science Framework, we have also created a website, www.LungInsight.ai, that will notify researchers on any updates to our project via newsletters and publications.
To make the data interoperable, we will share detailed metadata (workflows, vocabularies, processes, and standards) and the data will be shared in preservation friendly formats as detailed above. To make the data will share it under a Creative Commons CC-BY-4.0 license.
6. Responsibilities and Resources
6a. Identify who will be responsible for managing this project’s data during and after the project and the major data management tasks for which they will be responsible.
This Team will consist of
|
Individual |
Position |
Role |
|
|
Dr. Arvid Mer |
NPA |
Investigator |
|
|
Dr. Majid Komeili |
Co-principal applicant |
Investigator |
|
|
Dr. Manoj Lalu |
Co-principal applicant |
Investigator |
|
|
Dr. Dean Fergusson |
Co-applicant |
Investigator |
|
|
Dr. Arya Rahgozar |
Collaborator |
||
|
Dr. Sean Gil |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Haibo Zhang |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Arnold Kristof |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Bernard Thebaud |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Claudia DosSantos |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Christian Lehmann |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Duncan Stewart |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Braedon McDonald |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Katey Rayner |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Eric Schmidt |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Julie Bastarache |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Patrica Rocco |
Collaborator |
Sharing ALI histology slides |
|
|
Dr. Forough Jahandideh |
Research Associate |
Project manager |
|
|
Eva Kuhar |
HQP |
Annotator, managing ALI slides data and metadata |
|
|
Zoe Fisk |
HQP |
Annotator, managing ALI slides data and metadata |
|
|
Amir Ebrahimi |
HQP |
AI model development, Managing AI data and metadata |
|
|
MohammadReza Zarei |
HQP |
AI explainability and implementation, Managing AI data and metadata |
|
|
Brian Dorus |
Research Assistant |
Data management |
The names and specific roles of highly qualified personnel (HQP) will be added to this DMP as they are onboarded to the project.
6b. How will responsibilities for managing data activities be handled if substantive changes occur in the personnel overseeing the project’s data, including a change of Principal Investigator?
If any staff or trainees leave the project prior to completion, a current staff or trainee will replace them (or new personnel will be hired, if required). New personnel will receive adequate training to ensure competency image processing and lung scoring. Training will be enabled through project specific SOPs as well as videos found on www.LungInsight.ai. Furthermore, the staff/trainee will go through an off-boarding protocol outlining all the data they have generated, update any SOPs created, provide the location materials purchased, location of all the tissue/samples collected, etc
If the project needs to be transferred to a new principal investigator, then all responsibilities outlined in this DMP will be transferred as well.
6c. What resources will you require to implement your data management plan? What do you estimate the overall cost for data management to be?
The costs of implementing this DMP will consist of:
- Hiring a dedicated staff membrane to execute the DMP
- Fees for keeping www.LungInsight.ai running
For data sharing, since we are using the Open Science Framework, there is no cost for uploading our data. Given that our imaging data and AI models will exceed the 50GB capacity of Open Science Framework, we will house original images in the Federated Research Data Repository. We will clearly link to this data from our project on the Open Science Framework.
7. Ethics and Legal Compliance
7a. If your research project includes sensitive data, how will you ensure that it is securely managed and accessible only to approved members of the project?
Not applicable as only non-sensitive laboratory animal data will be collected/generated in our study.
7b. If applicable, what strategies will you undertake to address secondary uses of sensitive data?
Not applicable as the ‘participants’ are lung histology images.
7c. How will you manage legal, ethical, and intellectual property issues?
To ensure discoverability and access, we will deposit all datasets and accompanying metadata on the Open Science Framework (OSF). Once uploaded, we will receive a Digital Object Identifier (DOI), which we will cross-reference with publications, funding acknowledgements, and our institutional affiliation. In parallel, we will disseminate results through conference talks and poster sessions. Beyond the OSF archive, we will maintain a project website (www.LungInsight.ai) to announce updates, such as newsletters and publications, to the research community. To support interoperability, we will publish rich, machine-readable metadata documenting workflows, controlled vocabularies, processes, and standards, and we will share files in preservation-friendly formats as specified above. All materials will be released under a Creative Commons Attribution 4.0 (CC BY 4.0) license.
Version History
|
DMP version number |
Date Issued |
Summary of Revisions |
|
DMP_LungInsight_v0.1_20250812 |
15-08-2025 |
Initial draft by Brian Dorus; with comments provided by Amir Ebrahimi, Eva Kuhar, and Manoj Lalu |
|
DMP_LungInsight_v0.2_20251006 |
06-10-2025 |
Initial comments addressed by Brian Dorus; second round of comments provided by Manoj Lalu |
|
DMP_LungInsight_v0.3_20251007 |
07-10-2025 |
Second round of comments addressed by Brian Dorus |
|
DMP_LungInsight_v0.4_20251007 |
07-10-2025 |
Comments provided by Forough Jahandideh and Zoe Fisk, and addressed by Brian Dorus |
|
DMP_LungInsight_v0.5_20251009 |
09-10-2025 |
Comments provided by Arvind Mer, Majid Komeili, Amir Ebrahimi, and Mohammad Reza Zarei and addressed by Brian Dorus |
|
DMP_LungInsight_v1.0_20251010 |
10-10-2025 |
Comments addressed by Brian, with this version posted on Lunginsight.ai |
|
DMP_LungInsight_v1.1_20260222 |
22-02-2026 |
Comments provided by all of BLUEPRINT research group |
|
DMP_LungInsight_v2.0_20260302 |
02-03-2026 |
Comments addressed by Brian, with this version posted on lunginsight.ai |